Epigenome and lifestyle-related diseases
Introduction
Our living environment and habits rewrite our epigenome after birth, changing our constitution and susceptibility to diseases. In recent years, it has become clear that living environment and habits affect not only the individual but alto future children. Our research group focuses on the epigenome, and aim to elucidate the mechanisms by which environmental factors regulate biological phenomena and disease onset, and to develop of new methods of preventing and treating lifestyle-related diseases.
Research on fat burning/storing constitution and epigenome
Genetic information consists of genome and epigenome. The sequence of genomic DNA inherited from
parents basically does not change, However, the epigenome is rewritten in life depending on
living environment and habits. Representative examples of the epigenome include DNA methylation
and the methylation and acetylation of histone proteins, which play a role in winding DNA.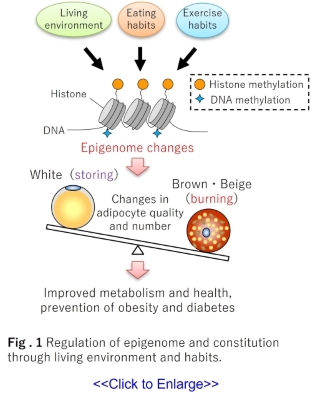
There are two types of adipocytes: white adipocytes, which store fat, and brown and beige
adipocytes which burn fat (1).
Brown and beige adipocytes actively metabolize sugar and fat and consume energy, so they are
attracting attention not only from the perspective of obesity prevention but also from the
perspective of health and beauty. Research to date has gradually revealed how living environment
and habits rewrite the epigenome of adipocytes and create a constitution that is more likely to
store or burn fat (Fig. 1)(2, 3).
Many people may not like the cold, but in fact, continuous exposure to cold rewrites the
epigenome of adipocytes, making the body more likely to burn fat (4-8).
In our laboratory, we are conducting further research into the mechanisms by which living
environment, eating habits, and exercise habits change the epigenome. We hope to use the
knowledge gained from our research to improve metabolism and create healthy bodies that are less
susceptible to disease.
Research on constitution and epigenome transmitted across generations
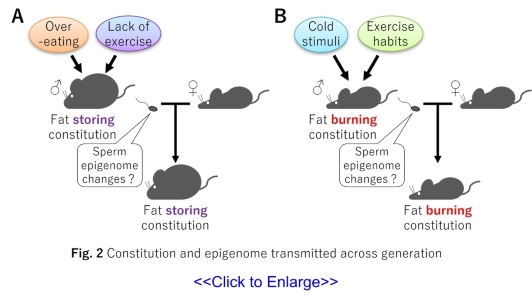
In recent years, it has become clear that a father’s living environment and habits affect not
only his own metabolism but also that of his future children. It has been reported that a
father’s bad lifestyle habits (unbalanced diet and lack of exercise) can be transmitted to his
children as a fat-storing constitution, increasing the risk of metabolic diseases(Fig. 2A) (9-12).
On the other hand, it has been reported that a father’s good lifestyle habit (cold exposure or
moderate exercise) can be transmitted to his children as a fat-burning constitution, decreasing
the risk of metabolic diseases(Fig. 2B)(11-13).
However, there are many unknows about the mechanism by which a father’s living environment and
habits are transmitted to his children as constitution. In our laboratory, we aim to clarify the
mechanism of transgenerational transmission of constitution from the perspective of epigenome by
analyzing DNA methylation and small RNAs in sperm. Through research into metabolic control
across generations, we aim to contribute to the health of the current and future people.
Research on the interplay between epigenome and epitranscriptome
In the central dogma of molecular biology, RNA is transcribed from DNA, and proteins are
translated from RNA.
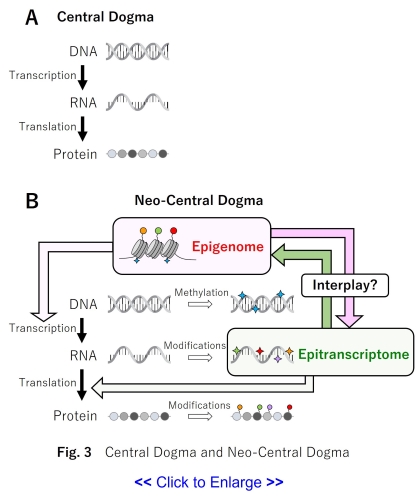 Through these processes, gene functions are expressed (Fig.
3A).
DNA methylation and chemical modifications of histone proteins are the epigenome, and these
epigenomes regulate the process of transcription from DNA to RNA.
On the other hand, RNA also undergoes various chemical modifications, and RNA modifications are
collectively called the epitranscriptome.
It has been found that RNA modifications regulate gene expression by controlling the
localization and stability of RNA, as well as the efficacy of translation into protein.
Conventionally, the epigenome and epitranscriptome were thought to regulate gene expression at
different layers.
However, in recent years, it has been suggested that epigenome controls epitranscriptome, and
epitranscriptome controls epigenome (14-17).
In our laboratory, we would like to approach the new concept of gene expression, the
“Neo-Central Dogma”, by analyzing reciprocal regulation of epigenome and epitranscriptome (Fig.
3B).
Through these processes, gene functions are expressed (Fig.
3A).
DNA methylation and chemical modifications of histone proteins are the epigenome, and these
epigenomes regulate the process of transcription from DNA to RNA.
On the other hand, RNA also undergoes various chemical modifications, and RNA modifications are
collectively called the epitranscriptome.
It has been found that RNA modifications regulate gene expression by controlling the
localization and stability of RNA, as well as the efficacy of translation into protein.
Conventionally, the epigenome and epitranscriptome were thought to regulate gene expression at
different layers.
However, in recent years, it has been suggested that epigenome controls epitranscriptome, and
epitranscriptome controls epigenome (14-17).
In our laboratory, we would like to approach the new concept of gene expression, the
“Neo-Central Dogma”, by analyzing reciprocal regulation of epigenome and epitranscriptome (Fig.
3B).
Searching for bioactive substances derived from microorganisms and chemical biology research
Introduction
Microorganisms have greatly enriched human life. In addition to their role in food and alcohol production, one of their most notable contributions is in the field of pharmaceutical development. The development of pharmaceuticals derived from microorganisms, which began with the discovery of antibiotics such as penicillin, has led to the creation of drugs with various biological activities, including anticancer drugs, cholesterol-lowering drugs, and immunosuppressants. Our research group focuses on discovering novel bioactive substances from the diverse secondary metabolites produced by microorganisms, aiming to contribute to the development of innovative pharmaceuticals.
Research on fibrinolysis-enhancing substances
Currently, ischemic heart disease ranks as the second leading cause of death in Japan, with
cerebrovascular diseases ranking fourth, so the treatment and prevention of thrombotic
disorders, which are major contributors to these diseases, are critically important worldwide.
Thrombolytic agents used in the treatment of thrombotic disorders enhance the fibrinolytic
system in blood vessels, leading to the degradation of formed thrombi. The fibrinolytic system
is an endogenous mechanism that maintains vascular homeostasis. It involves the conversion of
plasminogen, an inactive enzyme in the blood, into its active form, plasmin, through serine
proteases such as tissue-type plasminogen activator (tPA) and urokinase-type plasminogen
activator (uPA). Plasmin subsequently degrades fibrin, the main component of thrombi. Current
thrombolytic therapies use exogenous plasminogen activators such as tPA or uPA as enzyme-based
drugs in clinical settings. However, these therapies face significant challenges, including
short half-life in the bloodstream, the need for high-dose administration, systemic bleeding
tendencies, limitation to emergency use, and the inability for long-term application.
In the process of searching for fibrinolytic activity-enhancing substances aimed at developing
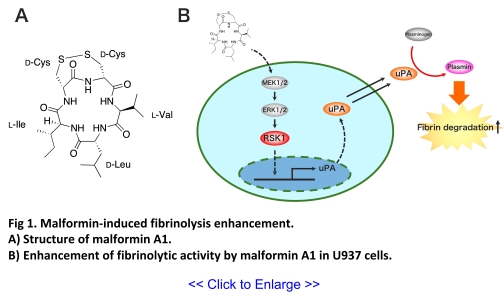
novel thrombolytic agents, we discovered malformin A1, a low-molecular-weight cyclic peptide
produced by filamentous fungi (Fig 1a) (1). Analysis
of mechanism of action revealed that
malformin enhances plasminogen activation via uPA produced by U937 monocytic cells, a model cell
line for the fibrinolysis research, at concentrations as low as a few μM (1, 2). Furthermore, it
promotes fibrinolytic activity by increasing uPA gene expression via the activation of the MAPK
pathway (3). These findings suggest that malformin enhances the
fibrinolytic system through a
mechanism of action distinct from that of existing enzyme-based drugs (Fig
1b). Currently, we
are investigating the effects of malformin on vascular endothelial cells, the primary
uPA-producing tissues within blood vessels.
Additionally, in collaboration with other institutions, we are conducting further screenings
for novel uPA activity-inducing substances. To date, these efforts have identified several
secondary metabolites of microbial origin.
Discovery of antibacterial substances
In recent years, the rise of drug-resistant bacteria that are unresponsive to antibiotics has
become a significant concern in many countries. A notable example is the 2004 outbreak of
methicillin-resistant Staphylococcus aureus (MRSA) and vancomycin-resistant enterococci (VRE) at
Akita University Hospital. Meanwhile, the development of new antibiotics has been on a declining
trend. One reason for this decline is that, compared to drugs for chronic diseases, antibiotics
are administered for shorter periods and therefore generate lower profits for pharmaceutical
companies. Consequently, many companies have withdrawn from antibiotic development, and very few
new antibiotics are being approved. Under these circumstances, there is an increasing need for
academia and public institutions to develop novel antibiotics.
To address this need, we conducted screening for new antibacterial substances using culture
extracts from microorganisms, such as filamentous fungi and actinomycetes, isolated from soil in
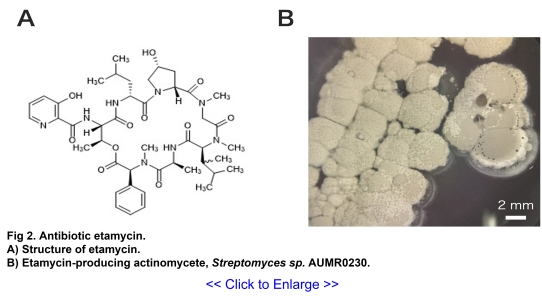
Akita Prefecture. This effort led to the discovery of etamycin A, a cyclic depsipeptide
belonging to group B streptogramin antibiotics, from a culture extract of actinomycete
Streptomyces sp. AUMR0230, which was isolated from river sediment in Semboku City (Fig 2)
(4, 5). Etamycin A exhibited potent antibacterial
activity against Gram-positive bacteria,
including methicillin-resistant Staphylococcus epidermidis (MRSE), a common pathogen in
hospital-acquired infections, with a minimum inhibitory concentration (MIC) of 1 μg/mL.
Furthermore, when combined with griseoviridin, a group A streptogramin antibiotic, etamycin A
demonstrated synergistic effects against MRSE. These findings indicate that etamycin A is an
effective streptogramin antibiotic against MRSE.
We are continuing to screen for antibacterial activity using microorganisms derived from Akita
Prefecture's soil. So far, we have identified several microorganisms capable of producing
antibacterial substances.
Discovery of brown adipose tissue-activating substances
In recent years, the adoption of Western dietary habits has led to an increasing prevalence of
obesity in Japan. Obesity is closely linked to lifestyle-related diseases such as diabetes,
hypertension, and dyslipidemia, and its progression raises the risk of arteriosclerosis.
Developing new therapeutic approaches for obesity has become an urgent challenge. Adipose tissue
is classified into white adipose tissue, which stores energy, and brown adipose tissue, which
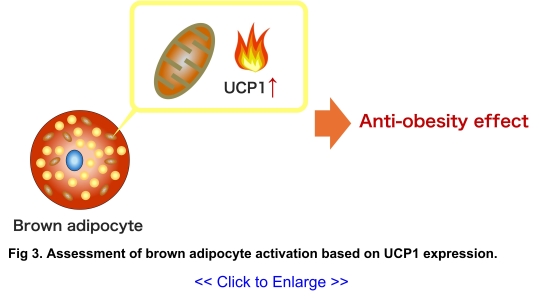
consumes energy. Activation of brown adipose tissue, which burns energy to generate heat, is
considered a promising anti-obesity strategy. We are currently establishing an evaluation system
for brown adipocyte activation based on the expression levels of mitochondrial uncoupling
protein 1 (UCP1), which is involved in thermogenesis in brown adipocytes (Fig
3). This
evaluation system is expected to facilitate screening efforts to discover novel anti-obesity
drugs that enhance the thermogenic capacity of brown adipocytes.
Cancer malignancy
Introduction
Cancer is one of the main causes of death worldwide. Tumor tissue may become malignant in certain cases. Malignant cancer cells invade into surrounding tissues and/or acquire metastatic property, which may lead to poor prognosis of patients. Our research group aims to elucidate the mechanism of cancer malignancy and to provide potential targets for cancer therapy.
Research on the interaction between cancer and the immune system
 Activation of the immune system is essential for cancer elimination, and research using mouse
tumor models has revealed that cancer malignancy is associated with immunosuppression, and that
cancer cells that adapt to an immunosuppressive environment promote malignancy through various
mechanisms.
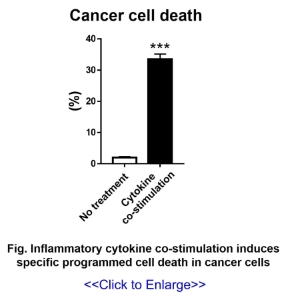
In our laboratory, we are studying the interaction between cancer and the immune system, and as
part of this, we have discovered specific cancer cell death due to cytokine stimulation (Fig)
and are currently analyzing its mechanism.
Activation of the immune system is essential for cancer elimination, and research using mouse
tumor models has revealed that cancer malignancy is associated with immunosuppression, and that
cancer cells that adapt to an immunosuppressive environment promote malignancy through various
mechanisms.
In our laboratory, we are studying the interaction between cancer and the immune system, and as
part of this, we have discovered specific cancer cell death due to cytokine stimulation (Fig)
and are currently analyzing its mechanism.
References
Epigenome and lifestyle-related diseases
- Matsumura Y, Osborne TF, Sakai J.
Epigenetic and environmental regulation of adipocyte function.
J Biochem (2022) 172, 9-16. - Matsumura Y, Ito R, Yajima A, Yamaguchi R, Tanaka T, Kawamura
T,
Magoori K, Abe Y, Uchida A, Yoneshiro T, Hirakawa H, Zhang J, Arai M, Yang C, Yang G,
Takahashi H, Fujihashi H, Nakaki R, Yamamoto S, Ota S, Tsutsumi S, Inoue SI, Kimura H, Wada
Y, Kodama T, Inagaki T, Osborne TF, Aburatani H, Node K, Sakai J.
H3K4/H3K9me3 Bivalent Chromatin Domains Targeted by Lineage-Specific DNA Methylation Pauses Adipocyte Differentiation.
Mol Cell (2015) 60, 584-596. - Matsumura Y, Nakaki R, Inagaki T, Yoshida A, Kano Y, Kimura H,
Tanaka T, Tsutsumi S, Nakao M, Doi T, Fukami K, Osborne TF, Kodama T, Aburatani H, Sakai
J.
Spatiotemporal dynamics of SETD5-containing NCoR-HDAC3 complex determines enhancer activation for adipogenesis.
Nat Commun (2021) 12, 7045. - Abe Y, Rozqie R, Matsumura Y, Kawamura T, Nakaki R, Tsurutani
Y,
Tanimura-Inagaki K, Shiono A, Magoori K, Nakamura K, Ogi S, Kajimura S, Kimura H, Tanaka T,
Fukami K, Osborne TF, Kodama T, Aburatani H, Inagaki T, Sakai J.
JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis.
Nat Commun (2015) 6, 7052. - Abe Y, Fujiwara Y, Takahashi H, Matsumura Y, Sawada T, Jiang S,
Nakaki R, Uchida A, Nagao N, Naito M, Kajimura S, Kimura H, Osborne TF, Aburatani H, Kodama
T, Inagaki T, Sakai J.
Histone demethylase JMJD1A coordinates acute and chronic adaptation to cold stress via thermogenic phospho-switch.
Nat Commun (2018) 9, 1566. - Takahashi H, Yang G, Yoneshiro T, Abe Y, Ito R, Yang C, Nakazono J,
Okamoto-Katsuyama M, Uchida A, Arai M, Jin H, Choi H, Tumenjargal M, Xie S, Zhang J, Sagae
H, Zhao Y, Yamaguchi R, Nomura Y, Shimizu Y, Yamada K, Yasuda S, Kimura H, Tanaka T, Wada Y,
Kodama T, Aburatani H, Zhu MS, Inagaki T, Osborne TF, Kawamura T, Ishihama Y,
Matsumura Y, Sakai J.
MYPT1-PP1β phosphatase negatively regulates both chromatin landscape and co-activator recruitment for beige adipogenesis.
Nat Commun (2022) 13, 5715. - Ito R, Xie S, Tumenjargal M, Sugahara Y, Yang C, Takahashi H, Arai M, Inoue SI,
Uchida A, Nakano K, Choi H, Yang G, Zhao Y, Yamaguchi R, Jin H, Sagae H, Wada Y, Tanaka T,
Kimura H, Kodama T, Aburatani H, Takeda K, Inagaki T, Osborne TF, Yoneshiro T,
Matsumura Y, Sakai J.
Mitochondrial biogenesis in white adipose tissue mediated by JMJD1A-PGC-1 axis limits age-related metabolic disease.
iScience (2024) 27, 109398. - Takahashi H, Ito R, Matsumura Y, Sakai J.
Environmental factor reversibly determines cellular identity through opposing Integrators that unify epigenetic and transcriptional pathways.
Bioessays (2024) 46, e2300084. - Sharma U, Conine CC, Shea JM, Boskovic A, Derr AG, Bing XY, Belleannee C,
Kucukural A, Serra RW, Sun F, Song L, Carone BR, Ricci EP, Li XZ, Fauquier L, Moore MJ,
Sullivan R, Mello CC, Garber M, Rando OJ.
Environmental factor reversibly determines cellular identity through opposing Integrators that unify epigenetic and transcriptional pathways.
Science (2016) 351, 391-396. - Chen Q, Yan M, Cao Z, Li X, Zhang Y, Shi J, Feng GH, Peng H, Zhang X, Zhang Y,
Qian J, Duan E, Zhai Q, Zhou Q.
Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder.
Science (2016) 351, 397-400. - Barrès R, Zierath JR.
The role of diet and exercise in the transgenerational epigenetic landscape of T2DM.
Nat Rev Endocrinol (2016) 12, 441-451. - Kusuyama J, Alves-Wagner AB, Makarewicz NS, Goodyear LJ.
Effects of maternal and paternal exercise on offspring metabolism.
Nat Metab (2020) 2, 858-872. - Sun W, Dong H, Becker AS, Dapito DH, Modica S, Grandl G, Opitz L, Efthymiou V,
Straub LG, Sarker G, Balaz M, Balazova L, Perdikari A, Kiehlmann E, Bacanovic S, Zellweger
C, Peleg-Raibstein D, Pelczar P, Reik W, Burger IA, von Meyenn F, Wolfrum C.
Cold-induced epigenetic programming of the sperm enhances brown adipose tissue activity in the offspring.
Nat Med (2018) 24, 1372-1383. - Huang H, Weng H, Zhou K, Wu T, Zhao BS, Sun M, Chen Z, Deng X, Xiao G, Auer F,
Klemm L, Wu H, Zuo Z, Qin X, Dong Y, Zhou Y, Qin H, Tao S, Du J, Liu J, Lu Z, Yin H,
Mesquita A, Yuan CL, Hu YC, Sun W, Su R, Dong L, Shen C, Li C, Qing Y, Jiang X, Wu X, Sun M,
Guan JL, Qu L, Wei M, Müschen M, Huang G, He C, Yang J, Chen J.
Histone H3 trimethylation at lysine 36 guides m6A RNA modification co-transcriptionally.
Nature (2019) 567, 414-419. - Murakami S, Jaffrey SR.
Hidden codes in mRNA: Control of gene expression by m(6)A.
Mol Cell (2022) 82, 2236-2251. - Kan RL, Chen J, Sallam T.
Crosstalk between epitranscriptomic and epigenetic mechanisms in gene regulation.
Trends Genet (2022) 38, 182-193. - Matsumura Y, Wei FY, Sakai J.
Epitranscriptomics in metabolic disease.
Nat Metab (2023) 5, 370-384.
Searching for bioactive substances derived from microorganisms and chemical biology research
- Koizumi Y, Hasumi K.
Enhancement of fibrinolytic activity of U937 cells by malformin A1.
J Antibiot (2002) 55, 78-82. - Koizumi Y, Fukudome H, Hasumi K.
Fibrinolytic activation promoted by the cyclopentapeptide malformin: involvement of cytoskeletal reorganization.
Biol Pharm Bull (2011) 34, 1426-1431. - Koizumi Y, Nagai K, Gao L, Koyota S, Yamaguchi T, Natsui M,
Imai Y, Hasumi K, Sugiyama T, Kuba K.
Involvement of RSK1 activation in malformin-enhanced cellular fibrinolytic activity.
Sci Rep (2018) 8, 5472. - Heinemann B, Gourevitch A, Lein J, Johnson DL, Kaplan MA, Vanas D, Hooper
IR.
Etamycin, a new antibiotic.
Antibiot Annu (1954) 2, 728-732. - Sheehan JC, Zachau HG, Lawson WB.
The structure of etamycin.
J Am Chem Soc (1958) 80, 3349-3355.
